|
 |
 |
 |
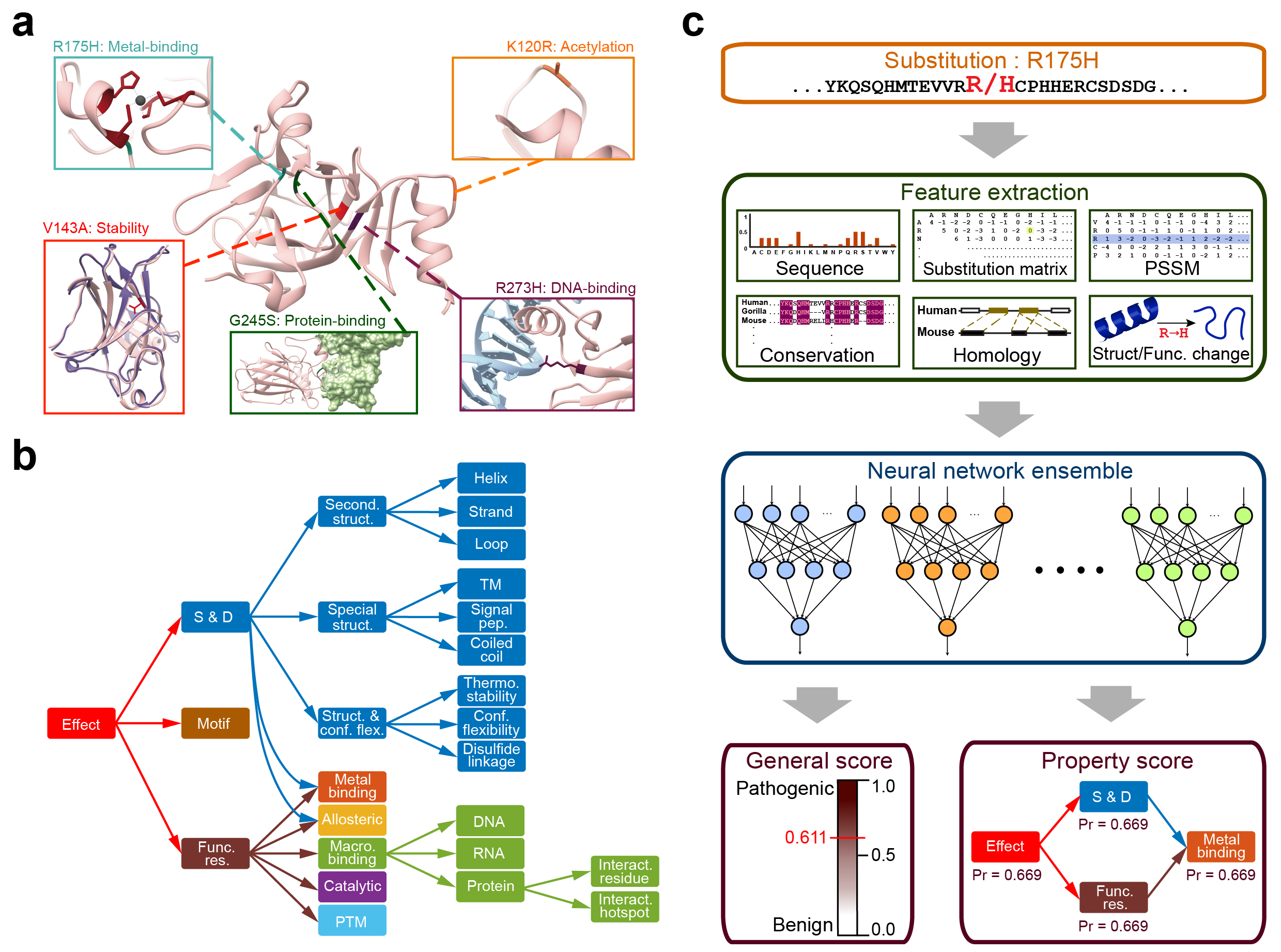
Predict the pathogenicity of amino acid substitutions and their molecular mechanisms
MutPred2 is a standalone and web application developed to classify amino acid substitutions as pathogenic or benign in human. In addition, it predicts their impact on over 50 different protein properties and, thus, enables the inference of molecular mechanisms of pathogenicity.
Predict here Download now Learn more
The MutPred2 paper can be found here. If you find MutPred2 useful in your work, please cite:
Pejaver V, Urresti J, Lugo-Martinez J, Pagel KA, Lin GN, Nam H, Mort M, Cooper DN, Sebat J, Iakoucheva LM, Mooney SD, Radivojac P. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 11, 5918 (2020)
The precomputed predictions from the original MutPred algorithm can be downloaded here. They can also be downloaded in dbNSFP.
For non-frameshift insertions and deletions, please use our new standalone and web application MutPred-Indel here.
For frameshift and stop-gain variants, please use our new standalone and web application MutPred-LOF here.
For splice variants, we have developed MutPred Splice here.